Our group works with collaborators in CALES and throughout the University to help implement computationally-intensive components of grant-funded projects. Click on the cards to read more about our current and past funded collaborations, or reach out if you are interested in working together!
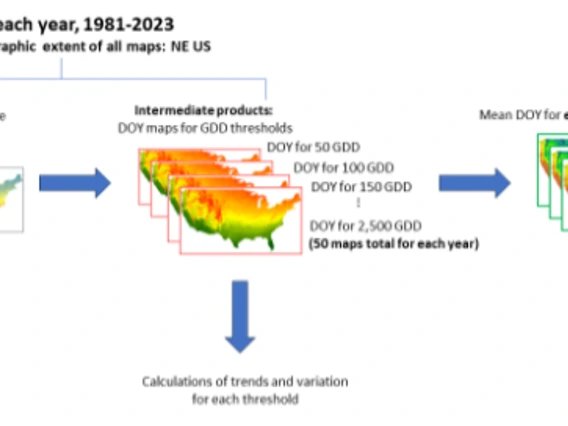
“The CCT Data Science Team has been incredibly helpful to the USA-NPN team, offering trustworthy analytical support and guidance. We have really benefited from their expertise in calculating temporal trends using raster datasets.”
- Dr. Theresa Crimmins, NPN Director & SNRE Associate Professor
“CCT Data Science has been critical in the modernization of AZMet (Arizona Meteorological Network) by setting up our GitHub organization, developing the `azmetr` package for programmatic access to our API in R, and building a data QA/QC dashboard with automated alert features. Their work now supports AZMet in day-to-day operations and sets the stage for further progress. More importantly, their work helps AZMet provide better service to its wide range of stakeholders, from individual growers to government agencies.”
- Dr. Jeremy Weiss, AZMet Program Manager, Cooperative Extension